User:StephanVCornell/Antimicrobial resistance
Mechanisms and organisms[edit]
Bacteria[edit]
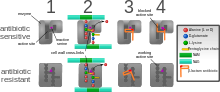
The five main mechanisms by which bacteria exhibit resistance to antibiotics are:
- Drug inactivation or modification: for example, enzymatic deactivation of penicillin G in some penicillin-resistant bacteria through the production of β-lactamases. Drugs may also be chemically modified through the addition of functional groups by transferase enzymes; for example, acetylation, phosphorylation, or adenylation are common resistance mechanisms to aminoglycosides. Acetylation is the most widely used mechanism and can affect a number of drug classes.[1][2]: 6–8
- Alteration of target- or binding site: for example, alteration of PBP—the binding target site of penicillins—in MRSA and other penicillin-resistant bacteria. Another protective mechanism found among bacterial species is ribosomal protection proteins. These proteins protect the bacterial cell from antibiotics that target the cell's ribosomes to inhibit protein synthesis. The mechanism involves the binding of the ribosomal protection proteins to the ribosomes of the bacterial cell, which in turn changes its conformational shape. This allows the ribosomes to continue synthesizing proteins essential to the cell while preventing antibiotics from binding to the ribosome to inhibit protein synthesis.[3]
- Alteration of metabolic pathway: for example, some sulfonamide-resistant bacteria do not require para-aminobenzoic acid (PABA), an important precursor for the synthesis of folic acid and nucleic acids in bacteria inhibited by sulfonamides, instead, like mammalian cells, they turn to using preformed folic acid.[4]
- Reduced drug accumulation: by decreasing drug permeability or increasing active efflux (pumping out) of the drugs across the cell surface[5] These pumps within the cellular membrane of certain bacterial species are used to pump antibiotics out of the cell before they are able to do any damage. They are often activated by a specific substrate associated with an antibiotic,[6] as in fluoroquinolone resistance.[7]
- Ribosome splitting and recycling: for example, drug-mediated stalling of the ribosome by lincomycin and erythromycin unstalled by a heat shock protein found in Listeria monocytogenes, which is a homologue of HflX from other bacteria. Liberation of the ribosome from the drug allows further translation and consequent resistance to the drug.[8]
Recent Developments:[edit]
- Cross-resistance: Exposure to one antibiotic can confer resistance to others, a critical factor in managing treatment regimens.
- Microgravity impacts on resistance: Studies have shown that non-pathogenic strains of E. coli can develop antibiotic resistance under simulated microgravity conditions, suggesting that the physical environment plays a role in resistance dynamics.
- Carbapenemases: The spread of carbapenemase-producing organisms, like those producing New Delhi metallo-beta-lactamase 1 (NDM-1), continues to be a significant challenge. These enzymes confer resistance to a broad spectrum of beta-lactam antibiotics and are primarily found in gram-negative bacteria such as E. coli and Klebsiella pneumoniae.
Impact of Resistance: The pathogens causing the most deaths associated with resistance include Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa. In 2019, these organisms were linked to approximately 929,000 deaths due to resistance and contributed to 3.57 million deaths associated with infections.
| This is the sandbox page where you will draft your initial Wikipedia contribution.
If you're starting a new article, you can develop it here until it's ready to go live. If you're working on improvements to an existing article, copy only one section at a time of the article to this sandbox to work on, and be sure to use an edit summary linking to the article you copied from. Do not copy over the entire article. You can find additional instructions here. Remember to save your work regularly using the "Publish page" button. (It just means 'save'; it will still be in the sandbox.) You can add bold formatting to your additions to differentiate them from existing content. |
References[edit]
- ^ Reygaert WC (2018). "An overview of the antimicrobial resistance mechanisms of bacteria". AIMS Microbiology. 4 (3): 482–501. doi:10.3934/microbiol.2018.3.482. PMC 6604941. PMID 31294229.
- ^ Baylay AJ, Piddock LJ, Webber MA (14 March 2019). "Molecular Mechanisms of Antibiotic Resistance – Part I". Bacterial Resistance to Antibiotics – from Molecules to Man: 1–26. doi:10.1002/9781119593522.ch1. ISBN 978-1-119-94077-7. S2CID 202806156.
- ^ Connell SR, Tracz DM, Nierhaus KH, Taylor DE (December 2003). "Ribosomal protection proteins and their mechanism of tetracycline resistance". Antimicrobial Agents and Chemotherapy. 47 (12): 3675–81. doi:10.1128/AAC.47.12.3675-3681.2003. PMC 296194. PMID 14638464.
- ^ Henry RJ (December 1943). "The Mode of Action of Sulfonamides". Bacteriological Reviews. 7 (4): 175–262. doi:10.1128/MMBR.7.4.175-262.1943. PMC 440870. PMID 16350088.
- ^ Li XZ, Nikaido H (August 2009). "Efflux-mediated drug resistance in bacteria: an update". Drugs. 69 (12): 1555–623. doi:10.2165/11317030-000000000-00000. PMC 2847397. PMID 19678712.
- ^ Aminov RI, Mackie RI (June 2007). "Evolution and ecology of antibiotic resistance genes". FEMS Microbiology Letters. 271 (2): 147–61. doi:10.1111/j.1574-6968.2007.00757.x. PMID 17490428.
- ^ Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T (July 1998). "NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli". Antimicrobial Agents and Chemotherapy. 42 (7): 1778–82. doi:10.1128/AAC.42.7.1778. PMC 105682. PMID 9661020.
- ^ Duval M, Dar D, Carvalho F, Rocha EP, Sorek R, Cossart P (December 2018). "HflXr, a homolog of a ribosome-splitting factor, mediates antibiotic resistance". Proceedings of the National Academy of Sciences of the United States of America. 115 (52): 13359–13364. Bibcode:2018PNAS..11513359D. doi:10.1073/pnas.1810555115. PMC 6310831. PMID 30545912.
