User:Mrdevgene/sandbox
H3K4me3[edit]
H3K4me3 is an epigenetic chemical modification involved in the regulation of gene expression.[1] The name denotes the addition of three methyl groups (trimethylation) to the lysine 4 on the histone H3 protein. H3 is used to package DNA in eukaryotic cells (including human cells). Modifications to histone proteins alter the accessibility of genes for transcription. H3K4me3 is commonly associated with transcription activation of nearby genes.[2] H3K4 trimethylation promotes gene expression through chromatin remodelling by the NURF complex.[3] This makes the DNA in the chromatin more accessible for transcription factors, allowing the genes to be transcribed and expressed in the cell. More specifically, H3K4me3 is found to positively regulate transcription by bringing histone acetylases and nucleosome remodelling enzymes (NURF).[4] H3K4me3 also plays an important role in the genetic regulation of stem cell potency and lineage.[5] This is because this histone modification is more-so found in areas of the DNA that are associated with development and establishing cell identity.[6]
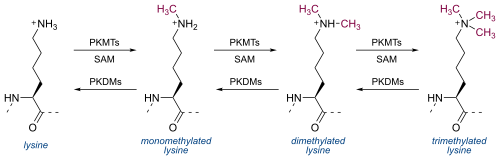
Lysine Methylation[edit]
The H3K4me3 modification is catalyzed by a lysine-specific histone methyltransferase (HMT), transferring three methyl groups to histone H3.[7] The methyltransferase complexes associated with H3K4me3 contains the protein WDR5, which consists of the WD40 repeat protein motif.[8] WDR5 associates specifically with dimethylated H3K4, which allows further methyltransferase activity, resulting in the creation and readout of the H3K4me3 modification.[9] WDR5 activity has been shown to be required for developmental genes, such as Hox genes, that are regulated by histone methylation.[8]
Epigenetic marker[edit]
H3K4me3 is one of the least abundant histone modifications; however, it is highly enriched at active promoters near transcription start sites (TSS)[10], and positively correlated with transcription. H3K4me3 is used as a histone code or histone mark in epigenetic studies (usually identified through chromatin immunoprecipitation) to identify active gene promoters. In fact, H3K4me3, as an epigenetic marker, has cancer researchers conclude that the treatment of a traditional Chinese medicine employs this histone modification at cancer-related gene loci.[11]
Role in stem cells and embryogenesis[edit]
Regulation of gene expression through H3K4me3 plays a significant role in stem cell fate determination and early embryo development. Pluripotent cells have distinctive patterns of methylation that can be identified through ChIP-sequencing. This is important in the development of induced pluripotent stem cells. A way of finding indicators of successful pluripotent induction is through comparing the epigenetic pattern to that of embryonic stem cells.[12]
In bivalent chromatin, H3K4me3 is co-localized with the repressive modification H3K27me3 to control gene regulation. H3K4me3 in embryonic cells is part of a bivalent chromatin system, in which regions of DNA are simultaneously marked with activating and repressing histone methylations.[13] This is believed to allow for a flexible system of gene expression, in which genes are primarily repressed, but may be expressed quickly due to H3K4me3 as the cell progresses through development.[5] These regions tend to coincide with transcription factor genes expressed at low levels. Some of these factors, such as the Hox genes, are essential to control development and cellular differentiation during embryogenesis.[3][5]
DNA repair[edit]
H3K4me3 is present at sites of DNA double-strand breaks where it promotes repair by the non-homologous end joining pathway. It has been implicated that the binding of H3K4me3 is necessary for the function of genes such as inhibitor of growth protein 1 (ING1), which act as a tumor suppressors and enact DNA repair mechanisms.[14]
When DNA damage occurs, DNA damage signalling and repair begins as a result of the modification of histones within the chromatin. Mechanistically, the demethylation of H3K4me3 is used required for specific protein binding and recruitment to DNA damage.[15]
 | This is a user sandbox of Mrdevgene. You can use it for testing or practicing edits. This is not the sandbox where you should draft your assigned article for a dashboard.wikiedu.org course. To find the right sandbox for your assignment, visit your Dashboard course page and follow the Sandbox Draft link for your assigned article in the My Articles section. |
- ^ Sims, Robert J.; Nishioka, Kenichi; Reinberg, Danny (November 2003). "Histone lysine methylation: a signature for chromatin function". Trends in genetics: TIG. 19 (11): 629–639. doi:10.1016/j.tig.2003.09.007. ISSN 0168-9525. PMID 14585615.
- ^ Wang, Feng; Tang, Zhongqiong; Shao, Honglian; Guo, Jun; Tan, Tao; Dong, Yang; Lin, Lianbing (June 12, 2018). "Long noncoding RNA HOTTIP cooperates with CCCTC-binding factor to coordinate HOXA gene expression". Biochemical and Biophysical Research Communications. 500 (4): 852–859. doi:10.1016/j.bbrc.2018.04.173. ISSN 1090-2104. PMID 29698677.
- ^ a b Wysocka, Joanna; Swigut, Tomek; Xiao, Hua; Milne, Thomas A.; Kwon, So Yeon; Landry, Joe; Kauer, Monika; Tackett, Alan J.; Chait, Brian T.; Badenhorst, Paul; Wu, Carl (2006-07-06). "A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling". Nature. 442 (7098): 86–90. doi:10.1038/nature04815. ISSN 1476-4687. PMID 16728976.
- ^ Santos-Rosa, Helena; Schneider, Robert; Bernstein, Bradley E.; Karabetsou, Nickoletta; Morillon, Antonin; Weise, Christoph; Schreiber, Stuart L.; Mellor, Jane; Kouzarides, Tony (2003). "Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin". Molecular Cell. 12 (5): 1325–1332. doi:10.1016/s1097-2765(03)00438-6. ISSN 1097-2765. PMID 14636589.
- ^ a b c Bernstein, Bradley E.; Mikkelsen, Tarjei S.; Xie, Xiaohui; Kamal, Michael; Huebert, Dana J.; Cuff, James; Fry, Ben; Meissner, Alex; Wernig, Marius; Plath, Kathrin; Jaenisch, Rudolf (2006-04-21). "A bivalent chromatin structure marks key developmental genes in embryonic stem cells". Cell. 125 (2): 315–326. doi:10.1016/j.cell.2006.02.041. ISSN 0092-8674. PMID 16630819.
- ^ Bernstein, Bradley E.; Mikkelsen, Tarjei S.; Xie, Xiaohui; Kamal, Michael; Huebert, Dana J.; Cuff, James; Fry, Ben; Meissner, Alex; Wernig, Marius; Plath, Kathrin; Jaenisch, Rudolf (2006-04-21). "A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells". Cell. 125 (2): 315–326. doi:10.1016/j.cell.2006.02.041. ISSN 0092-8674. PMID 16630819.
- ^ Rice, Judd C.; Briggs, Scott D.; Ueberheide, Beatrix; Barber, Cynthia M.; Shabanowitz, Jeffrey; Hunt, Donald F.; Shinkai, Yoichi; Allis, C. David (December 2003). "Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains". Molecular Cell. 12 (6): 1591–1598. doi:10.1016/s1097-2765(03)00479-9. ISSN 1097-2765. PMID 14690610.
- ^ a b Wysocka, Joanna; Swigut, Tomek; Milne, Thomas A.; Dou, Yali; Zhang, Xin; Burlingame, Alma L.; Roeder, Robert G.; Brivanlou, Ali H.; Allis, C. David (2005-06-17). "WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development". Cell. 121 (6): 859–872. doi:10.1016/j.cell.2005.03.036. ISSN 0092-8674. PMID 15960974.
- ^ Ruthenburg, Alexander J.; Wang, Wooikoon; Graybosch, Daina M.; Li, Haitao; Allis, C. David; Patel, Dinshaw J.; Verdine, Gregory L. (August 2006). "Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex". Nature Structural & Molecular Biology. 13 (8): 704–712. doi:10.1038/nsmb1119. ISSN 1545-9993. PMC 4698793. PMID 16829959.
- ^ Liang, Gangning; Lin, Joy C. Y.; Wei, Vivian; Yoo, Christine; Cheng, Jonathan C.; Nguyen, Carvell T.; Weisenberger, Daniel J.; Egger, Gerda; Takai, Daiya; Gonzales, Felicidad A.; Jones, Peter A. (2004-05-11). "Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome". Proceedings of the National Academy of Sciences of the United States of America. 101 (19): 7357–7362. doi:10.1073/pnas.0401866101. ISSN 0027-8424. PMID 15123803.
- ^ Lu, Jun; Zhang, Xiaoli; Shen, Tingting; Ma, Chao; Wu, Jun; Kong, Hualei; Tian, Jing; Shao, Zhifeng; Zhao, Xiaodong; Xu, Ling (2016). "Epigenetic Profiling of H3K4Me3 Reveals Herbal Medicine Jinfukang-Induced Epigenetic Alteration Is Involved in Anti-Lung Cancer Activity". Evidence-Based Complementary and Alternative Medicine: eCAM. 2016: 7276161. doi:10.1155/2016/7276161. ISSN 1741-427X. PMC 4818803. PMID 27087825.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Tesar, Paul J.; Chenoweth, Josh G.; Brook, Frances A.; Davies, Timothy J.; Evans, Edward P.; Mack, David L.; Gardner, Richard L.; McKay, Ronald D. G. (2007-07-12). "New cell lines from mouse epiblast share defining features with human embryonic stem cells". Nature. 448 (7150): 196–199. doi:10.1038/nature05972. ISSN 1476-4687. PMID 17597760.
- ^ Vastenhouw, Nadine L.; Schier, Alexander F. (June 2012). "Bivalent histone modifications in early embryogenesis". Current Opinion in Cell Biology. 24 (3): 374–386. doi:10.1016/j.ceb.2012.03.009. ISSN 1879-0410. PMC 3372573. PMID 22513113.
- ^ Peña, P. V.; Hom, R. A.; Hung, T.; Lin, H.; Kuo, A. J.; Wong, R. P. C.; Subach, O. M.; Champagne, K. S.; Zhao, R.; Verkhusha, V. V.; Li, G. (2008-07-04). "Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor". Journal of Molecular Biology. 380 (2): 303–312. doi:10.1016/j.jmb.2008.04.061. ISSN 1089-8638. PMC 2576750. PMID 18533182.
- ^ Gong, Fade; Clouaire, Thomas; Aguirrebengoa, Marion; Legube, Gaëlle; Miller, Kyle M. (July 3, 2017). "Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair". The Journal of Cell Biology. 216 (7): 1959–1974. doi:10.1083/jcb.201611135. ISSN 1540-8140. PMC 5496618. PMID 28572115.
