Working example [ edit ] This working example is based on a JC69 genetic distance matrix computed from the 5S ribosomal RNA sequence alignment of five bacteria: Bacillus subtilis
a
{\displaystyle a}
Bacillus stearothermophilus
b
{\displaystyle b}
Lactobacillus viridescens
c
{\displaystyle c}
Acholeplasma modicum
d
{\displaystyle d}
Micrococcus luteus
e
{\displaystyle e}
[1] [2]
First step [ edit ] Let us assume that we have five elements
(
a
,
b
,
c
,
d
,
e
)
{\displaystyle (a,b,c,d,e)}
D
1
{\displaystyle D_{1}}
a
b
c
d
e
a
0
17
21
31
23
b
17
0
30
34
21
c
21
30
0
28
39
d
31
34
28
0
43
e
23
21
39
43
0
In this example,
D
1
(
a
,
b
)
=
17
{\displaystyle D_{1}(a,b)=17}
D
1
{\displaystyle D_{1}}
a
{\displaystyle a}
b
{\displaystyle b}
First branch length estimation Let
u
{\displaystyle u}
a
{\displaystyle a}
b
{\displaystyle b}
δ
(
a
,
u
)
=
δ
(
b
,
u
)
=
D
1
(
a
,
b
)
/
2
{\displaystyle \delta (a,u)=\delta (b,u)=D_{1}(a,b)/2}
a
{\displaystyle a}
b
{\displaystyle b}
u
{\displaystyle u}
ultrametricity hypothesis.
The branches joining
a
{\displaystyle a}
b
{\displaystyle b}
u
{\displaystyle u}
δ
(
a
,
u
)
=
δ
(
b
,
u
)
=
17
/
2
=
8.5
{\displaystyle \delta (a,u)=\delta (b,u)=17/2=8.5}
see the final dendrogram
First distance matrix update We then proceed to update the initial proximity matrix
D
1
{\displaystyle D_{1}}
D
2
{\displaystyle D_{2}}
a
{\displaystyle a}
b
{\displaystyle b}
D
2
{\displaystyle D_{2}}
maximum distance between each element of the first cluster
(
a
,
b
)
{\displaystyle (a,b)}
D
2
(
(
a
,
b
)
,
c
)
=
m
a
x
(
D
1
(
a
,
c
)
,
D
1
(
b
,
c
)
)
=
m
a
x
(
21
,
30
)
=
30
{\displaystyle D_{2}((a,b),c)=max(D_{1}(a,c),D_{1}(b,c))=max(21,30)=30}
D
2
(
(
a
,
b
)
,
d
)
=
m
a
x
(
D
1
(
a
,
d
)
,
D
1
(
b
,
d
)
)
=
m
a
x
(
31
,
34
)
=
34
{\displaystyle D_{2}((a,b),d)=max(D_{1}(a,d),D_{1}(b,d))=max(31,34)=34}
D
2
(
(
a
,
b
)
,
e
)
=
m
a
x
(
D
1
(
a
,
e
)
,
D
1
(
b
,
e
)
)
=
m
a
x
(
23
,
21
)
=
23
{\displaystyle D_{2}((a,b),e)=max(D_{1}(a,e),D_{1}(b,e))=max(23,21)=23}
Italicized values in
D
2
{\displaystyle D_{2}}
Second step [ edit ] We now reiterate the three previous steps, starting from the new distance matrix
D
2
{\displaystyle D_{2}}
(a,b)
c
d
e
(a,b)
0
30 34 23
c
30 0
28 39
d
34 28 0
43
e
23 39 43 0
Here,
D
2
(
(
a
,
b
)
,
e
)
=
23
{\displaystyle D_{2}((a,b),e)=23}
D
2
{\displaystyle D_{2}}
(
a
,
b
)
{\displaystyle (a,b)}
e
{\displaystyle e}
Second branch length estimation Let
v
{\displaystyle v}
(
a
,
b
)
{\displaystyle (a,b)}
e
{\displaystyle e}
a
{\displaystyle a}
b
{\displaystyle b}
v
{\displaystyle v}
e
{\displaystyle e}
v
{\displaystyle v}
δ
(
a
,
v
)
=
δ
(
b
,
v
)
=
δ
(
e
,
v
)
=
23
/
2
=
11.5
{\displaystyle \delta (a,v)=\delta (b,v)=\delta (e,v)=23/2=11.5}
We deduce the missing branch length:
δ
(
u
,
v
)
=
δ
(
e
,
v
)
−
δ
(
a
,
u
)
=
δ
(
e
,
v
)
−
δ
(
b
,
u
)
=
11.5
−
8.5
=
3
{\displaystyle \delta (u,v)=\delta (e,v)-\delta (a,u)=\delta (e,v)-\delta (b,u)=11.5-8.5=3}
see the final dendrogram
Second distance matrix update We then proceed to update the
D
2
{\displaystyle D_{2}}
D
3
{\displaystyle D_{3}}
(
a
,
b
)
{\displaystyle (a,b)}
e
{\displaystyle e}
D
3
(
(
(
a
,
b
)
,
e
)
,
c
)
=
m
a
x
(
D
2
(
(
a
,
b
)
,
c
)
,
D
2
(
e
,
c
)
)
=
m
a
x
(
30
,
39
)
=
39
{\displaystyle D_{3}(((a,b),e),c)=max(D_{2}((a,b),c),D_{2}(e,c))=max(30,39)=39}
D
3
(
(
(
a
,
b
)
,
e
)
,
d
)
=
m
a
x
(
D
2
(
(
a
,
b
)
,
d
)
,
D
2
(
e
,
d
)
)
=
m
a
x
(
34
,
43
)
=
43
{\displaystyle D_{3}(((a,b),e),d)=max(D_{2}((a,b),d),D_{2}(e,d))=max(34,43)=43}
Third step [ edit ] We again reiterate the three previous steps, starting from the updated distance matrix
D
3
{\displaystyle D_{3}}
((a,b),e)
c
d
((a,b),e)
0
39 43
c
39 0
28
d
43 28 0
Here,
D
3
(
c
,
d
)
=
28
{\displaystyle D_{3}(c,d)=28}
D
3
{\displaystyle D_{3}}
c
{\displaystyle c}
d
{\displaystyle d}
Third branch length estimation Let
w
{\displaystyle w}
c
{\displaystyle c}
d
{\displaystyle d}
c
{\displaystyle c}
d
{\displaystyle d}
w
{\displaystyle w}
δ
(
c
,
w
)
=
δ
(
d
,
w
)
=
28
/
2
=
14
{\displaystyle \delta (c,w)=\delta (d,w)=28/2=14}
see the final dendrogram
Third distance matrix update There is a single entry to update:
D
4
(
(
c
,
d
)
,
(
(
a
,
b
)
,
e
)
)
=
(
D
3
(
c
,
(
(
a
,
b
)
,
e
)
)
,
D
3
(
d
,
(
(
a
,
b
)
,
e
)
)
)
=
(
39
,
43
)
=
43
{\displaystyle D_{4}((c,d),((a,b),e))=(D_{3}(c,((a,b),e)),D_{3}(d,((a,b),e)))=(39,43)=43}
Final step [ edit ] The final
D
4
{\displaystyle D_{4}}
((a,b),e)
(c,d)
((a,b),e)
0
43
(c,d)
43 0
So we join clusters
(
(
a
,
b
)
,
e
)
{\displaystyle ((a,b),e)}
(
c
,
d
)
{\displaystyle (c,d)}
Let
r
{\displaystyle r}
(
(
a
,
b
)
,
e
)
{\displaystyle ((a,b),e)}
(
c
,
d
)
{\displaystyle (c,d)}
(
(
a
,
b
)
,
e
)
{\displaystyle ((a,b),e)}
(
c
,
d
)
{\displaystyle (c,d)}
r
{\displaystyle r}
δ
(
(
(
a
,
b
)
,
e
)
,
r
)
=
δ
(
(
c
,
d
)
,
r
)
=
43
/
2
=
21.5
{\displaystyle \delta (((a,b),e),r)=\delta ((c,d),r)=43/2=21.5}
We deduce the two remaining branch lengths:
δ
(
v
,
r
)
=
δ
(
(
(
a
,
b
)
,
e
)
,
r
)
−
δ
(
e
,
v
)
=
21.5
−
11.5
=
10
{\displaystyle \delta (v,r)=\delta (((a,b),e),r)-\delta (e,v)=21.5-11.5=10}
δ
(
w
,
r
)
=
δ
(
(
c
,
d
)
,
r
)
−
δ
(
c
,
w
)
=
21.5
−
14
=
7.5
{\displaystyle \delta (w,r)=\delta ((c,d),r)-\delta (c,w)=21.5-14=7.5}
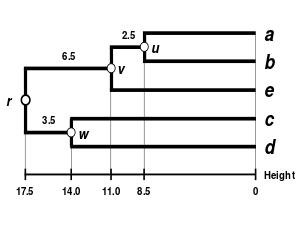
The complete-linkage dendrogram [ edit ]
WPGMA Dendrogram 5S data The dendrogram is now complete. It is ultrametric because all tips (
a
{\displaystyle a}
e
{\displaystyle e}
r
{\displaystyle r}
δ
(
a
,
r
)
=
δ
(
b
,
r
)
=
δ
(
e
,
r
)
=
δ
(
c
,
r
)
=
δ
(
d
,
r
)
=
21.5
{\displaystyle \delta (a,r)=\delta (b,r)=\delta (e,r)=\delta (c,r)=\delta (d,r)=21.5}
The dendrogram is therefore rooted by
r
{\displaystyle r}
Comparison of clustering methods [ edit ]
Comparison of dendrograms obtained under different clustering methods from the same distance matrix.
Single-linkage clustering
Complete-linkage clustering
WPGMA
UPGMA