Draft:SimPlot++
| Submission declined on 7 April 2024 by Sohom Datta (talk). This draft's references do not show that the subject qualifies for a Wikipedia article. In summary, the draft needs multiple published sources that are:
Where to get help
How to improve a draft
You can also browse Wikipedia:Featured articles and Wikipedia:Good articles to find examples of Wikipedia's best writing on topics similar to your proposed article. Improving your odds of a speedy review To improve your odds of a faster review, tag your draft with relevant WikiProject tags using the button below. This will let reviewers know a new draft has been submitted in their area of interest. For instance, if you wrote about a female astronomer, you would want to add the Biography, Astronomy, and Women scientists tags. Editor resources
|  |
| Submission declined on 8 November 2023 by Vanderwaalforces (talk). This submission does not appear to be written in the formal tone expected of an encyclopedia article. Entries should be written from a neutral point of view, and should refer to a range of independent, reliable, published sources. Please rewrite your submission in a more encyclopedic format. Please make sure to avoid peacock terms that promote the subject. This submission appears to read more like an advertisement than an entry in an encyclopedia. Encyclopedia articles need to be written from a neutral point of view, and should refer to a range of independent, reliable, published sources, not just to materials produced by the creator of the subject being discussed. This is important so that the article can meet Wikipedia's verifiability policy and the notability of the subject can be established. If you still feel that this subject is worthy of inclusion in Wikipedia, please rewrite your submission to comply with these policies. |  |
| Submission declined on 20 September 2023 by OlifanofmrTennant (talk). This submission appears to read more like an advertisement than an entry in an encyclopedia. Encyclopedia articles need to be written from a neutral point of view, and should refer to a range of independent, reliable, published sources, not just to materials produced by the creator of the subject being discussed. This is important so that the article can meet Wikipedia's verifiability policy and the notability of the subject can be established. If you still feel that this subject is worthy of inclusion in Wikipedia, please rewrite your submission to comply with these policies. |  |
 Comment: After doing a skim through a bunch of sources, I'm unable to verify the source-to-text integrity of the article. Please rewrite the article so that each and every citation mentions SimPlot++ and talks about the tool in the context. Randomly linking to sources that discuss the context without mentioning the tool will lead to your draft being declined again. Sohom (talk) 23:15, 7 April 2024 (UTC)
Comment: After doing a skim through a bunch of sources, I'm unable to verify the source-to-text integrity of the article. Please rewrite the article so that each and every citation mentions SimPlot++ and talks about the tool in the context. Randomly linking to sources that discuss the context without mentioning the tool will lead to your draft being declined again. Sohom (talk) 23:15, 7 April 2024 (UTC)
SimPlot++.[1] is a free multi-platform application which allows the users to generate publication-quality sequence similarity plots. This open-source program was developed at the department of Computer Science of the Université du Québec à Montréal. SimPlot++ incudes 20 amino acid and 63 nucleotide evolutionary models. It allows the users to draw high-quality sequence similarity plots, identify recombination events (χ2 [2], Neighbour Similarity Score [3], Pairwise Homoplasy Index [4] and Proportion recombinations tests are available), and visualize and investigate sequence similarity networks. Improved versions of the SimPlot, FindSites and BootScan tools (originally proposed in the SimPlot software) [5] have been implemented and made available in SimPlot++.
The Windows, Mac OS and Linux versions of the program can be freely downloaded from github. SimPlot++ is a new addition to other similarity plot software tools, including RIP [6], SimPlot [5], BLAST Genotyping tool [7], T-RECs [8] and RDP5 [9], which have been widely used in many bioinformatics and evolutionary studies.
General Use[edit]
Simplot++ takes as input a multiple sequence alignment of DNA or Amino acid sequences. The input sequences should be aligned and encoded using one of the following data formats: FASTA, Clustal, Nexus, Stockholm, PIR or PHYLIP.
The input sequences loaded into SimPlot++ should be separated in different groups by the user, based on their evolutionary resemblance. This will allow the program to create consensus sequences.[10] (i.e. group sequences). The consensus sequences will then be used to carry out all analyses available in SimPlot++. The minimum number of groups which should be created is two (to get full access to most of the program features)[1]
SimPlot analysis[edit]

The multiple sequence alignment (MSA)[5] (MSA) provided by the user is processed by means of a sliding window-based algorithm using as parameters a specified progress step and a sliding window size to carry out SimPlot analysis [5]. The portion of the MSA corresponding to the current location of the sliding window is used to compute an evolutionary distance matrix with respect to the selected evolutionary distance model. This matrix is processed by the program to generate a sequence similarity plot that shows similarity between all available individual and consensus sequences and the selected reference sequence.
To conduct the SimPlot analysis[1] one can use 63 DNA and 20 amino acid evolutionary models, activate a multiprocessing option to speed up the calculation process, or take advantage of Matplotlib-based sequence similarity graphs which can be easily modified and saved in a variety of formats[11]. SimPlot++ also offers a novel distance calculability diagnostic option to show for which sequence regions the distance calculation was impossible (if any).
BootScan analysis[edit]
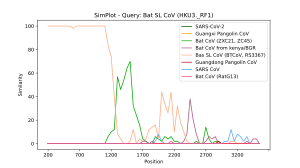
The BootScan analysis [12] available in SimPlot++ relies on the four following steps. The consensus sequence fragments covered by the current position of the sliding window (i.e. the sub-MSA corresponding to the current position of the sliding window) are bootstrapped n times. For each such a sub-MSA, the corresponding distance matrix is generated and a phylogeny[13][14] is constructed using either Neighbor joining[15] or UPGMA[16] algorithms. The number of trees in which the reference sequence is the nearest neighbor of each consensus sequence is calculated. The same variety of evolutionary distance models, the multiprocessing capability, and the similarity plot options as for SimPlot analysis are also available with Bootscan analysis. Importantly, the selected sequences must be aligned before carrying out the BootScan analysis.
FindSites[edit]
The FindSites analysis is usually carried out to detect eventual recombined sequence regions by using the Informative sites[17] method. Four sequences of interest are involved in the analysis. One of them is supposed to be a sequence resulting from recombination, two of them are supposed to be the parents of the first sequence, and the forth one plays the role of an outgroup. The Informative sites[17] method identifies the sites of interest as those at which two of the four sequences share the same nucleotides, while the other two share a different one.
Similarity Network[edit]

Sequence similarity networks[18] [19] can be used to represent similarity relationships by visualizing different similarity links between evolutionary sequences. In a sequence similarity network every sequence, or group of sequences, is represented by a single vertex (or network node). A given pair of vertices in such a network can be linked (or not linked) to each other by a branch depending on the value of the evolutionary distance between these sequences and the similarity threshold selected by the user. Both local (when only some sequence regions are considered) and global sequence similarity[1] can be represented using this kind of graphs. The similarity threshold can be changed to better visualize either local or global similarity connections. Moreover, a similarity network can correspond to a selected sequence region of a given MSA. This can help one analyze in greater details a specific gene or gene region of a given MSA. The networks produced by SimPlot++ can be saved in the png, svg or HTML formats. Importantly, the selected sequences must be aligned before carrying out the network analysis. It is worth noting that the sequence similarity network analysis is available in SimPlot++ only (it is not available in SimPlot[5]).
Recombination analysis[edit]
Several statistical tests for identifying recombination (i.e. mosaic sequence regions[20]) have been included in SimPlot++. Some of them come from the PhiPack[4] package. These tests include the Phi[4] (Pairwise Homoplasy Index) test, the Phi-profile[4] (Pairwise Homoplasy Index-profile) test, the Neighbour Similarity Score (NSS) test[3] and the Maximum χ2 test[2]. These tests can be conducted for both ungrouped (i.e. original) and grouped sequences. Importantly, the selected sequences must be aligned before carrying out the recombination analysis. In addition, a novel fast Proportion test can be also carried out to detect recombination events in large genomic sequences. Once again, the described options of recombination analysis are available in SimPlot++ only (they are not available in SimPlot[5]). Other existing programs for detecting recombination include: TOPALi [21], RAT [22], T-RECs [8] and RDP5 [9]
References[edit]
- ^ a b c d Samson, Stéphane; Lord, Étienne; Makarenkov, Vladimir (April 2022). "SimPlot++: a Python application for representing sequence similarity and detecting recombination". Bioinformatics. 38 (11): 3118–3120. arXiv:2112.09755. doi:10.1093/bioinformatics/btac287. PMID 35451456.
- ^ a b Smith, JohnMaynard (February 1992). "Analyzing the mosaic structure of genes". Journal of Molecular Evolution. 34 (2): 126–129. Bibcode:1992JMolE..34..126S. doi:10.1007/BF00182389. PMID 1556748. S2CID 5111557.
- ^ a b Jakobsen, Ingrid B.; Easteal, Simon (1996). "A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences". Bioinformatics. 12 (4): 291–295. doi:10.1093/bioinformatics/12.4.291. PMID 8902355.
- ^ a b c d Bruen, Trevor C; Philippe, Hervé; Bryant, David (1 April 2006). "A Simple and Robust Statistical Test for Detecting the Presence of Recombination". Genetics. 172 (4): 2665–2681. doi:10.1534/genetics.105.048975. PMC 1456386. PMID 16489234.
- ^ a b c d e f Lole, Kavita S.; Bollinger, Robert C.; Paranjape, Ramesh S.; Gadkari, Deepak; Kulkarni, Smita S.; Novak, Nicole G.; Ingersoll, Roxann; Sheppard, Haynes W.; Ray, Stuart C. (January 1999). "Full-Length Human Immunodeficiency Virus Type 1 Genomes from Subtype C-Infected Seroconverters in India, with Evidence of Intersubtype Recombination". Journal of Virology. 73 (1): 152–160. doi:10.1128/jvi.73.1.152-160.1999. PMC 103818. PMID 9847317.
- ^ Siepel, Adam C.; Halpern, Aaron L.; Macken, Catherine; Korber, Bette T.m. (November 1995). "A Computer Program Designed to Screen Rapidly for HIV Type 1 Intersubtype Recombinant Sequences". AIDS Research and Human Retroviruses. 11 (11): 1413–1416. doi:10.1089/aid.1995.11.1413. PMID 8573400.
- ^ "Genotyping". www.ncbi.nlm.nih.gov. Retrieved 8 November 2023.
- ^ a b Tsimpidis, Michail; Bachoumis, Georgios; Mimouli, Kalliopi; Kyriakopoulou, Zaharoula; Robertson, David L.; Markoulatos, Panayotis; Amoutzias, Grigoris D. (December 2017). "T-RECs: rapid and large-scale detection of recombination events among different evolutionary lineages of viral genomes". BMC Bioinformatics. 18 (1): 13. doi:10.1186/s12859-016-1420-z. PMC 5216575. PMID 28056784.
- ^ a b Martin, Darren P; Varsani, Arvind; Roumagnac, Philippe; Botha, Gerrit; Maslamoney, Suresh; Schwab, Tiana; Kelz, Zena; Kumar, Venkatesh; Murrell, Ben (12 April 2020). "RDP5: a computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets". Virus Evolution. 7 (1): veaa087. doi:10.1093/ve/veaa087. PMC 8062008. PMID 33936774.
- ^ Schneider, Thomas D. (2002). "Consensus Sequence Zen". Applied Bioinformatics. 1 (3): 111–119. ISSN 1175-5636. PMC 1852464. PMID 15130839.
- ^ Hunter, John D. (2007). "Matplotlib: A 2D Graphics Environment". Computing in Science & Engineering. 9 (3): 90–95. Bibcode:2007CSE.....9...90H. doi:10.1109/MCSE.2007.55. S2CID 37016120.
- ^ Salminen, Mika O.; Carr, Jean K.; Burke, Donald S.; McCUTCHAN, Francine E. (1 November 1995). "Identification of Breakpoints in Intergenotypic Recombinants of HIV Type 1 by Bootscanning". AIDS Research and Human Retroviruses. 11 (11): 1423–1425. doi:10.1089/aid.1995.11.1423. ISSN 0889-2229. PMID 8573403.
- ^ Felsenstein, Joseph (2004). Inferring phylogenies. Sunderland, MA: Sinauer associates.
- ^ Penny, David (1 August 2004). "Inferring Phylogenies.—Joseph Felsenstein. 2003. Sinauer Associates, Sunderland, Massachusetts". Systematic Biology. 53 (4): 669–670. doi:10.1080/10635150490468530.
- ^ Saitou, N.; Nei, M. (July 1987). "The neighbor-joining method: a new method for reconstructing phylogenetic trees". Molecular Biology and Evolution. 4 (4): 406–425. doi:10.1093/oxfordjournals.molbev.a040454. ISSN 0737-4038. PMID 3447015.
- ^ Sokal, Michener (1958). "A statistical method for evaluating systematic relationships". University of Kansas Science Bulletin. 38: 1409–1438.
- ^ a b Robertson, David L.; Hahn, Beatrice H.; Sharp, Paul M. (1 March 1995). "Recombination in AIDS viruses". Journal of Molecular Evolution. 40 (3): 249–259. Bibcode:1995JMolE..40..249R. doi:10.1007/BF00163230. ISSN 1432-1432. PMID 7723052. S2CID 19728830.
- ^ Xing, Henry; Kembel, Steven W; Makarenkov, Vladimir (1 May 2020). "Transfer index, NetUniFrac and some useful shortest path-based distances for community analysis in sequence similarity networks". Bioinformatics. 36 (9): 2740–2749. doi:10.1093/bioinformatics/btaa043. PMID 31971565.
- ^ Bapteste, Eric; van Iersel, Leo; Janke, Axel; Kelchner, Scot; Kelk, Steven; McInerney, James O.; Morrison, David A.; Nakhleh, Luay; Steel, Mike; Stougie, Leen; Whitfield, James (August 2013). "Networks: expanding evolutionary thinking". Trends in Genetics. 29 (8): 439–441. doi:10.1016/j.tig.2013.05.007. PMID 23764187. S2CID 8996825.
- ^ Forsberg, Lars A.; Gisselsson, David; Dumanski, Jan P. (February 2017). "Mosaicism in health and disease — clones picking up speed". Nature Reviews Genetics. 18 (2): 128–142. doi:10.1038/nrg.2016.145. PMID 27941868. S2CID 44092954.
- ^ Milne, Iain; Wright, Frank; Rowe, Glenn; Marshall, David F.; Husmeier, Dirk; McGuire, Gràinne (22 July 2004). "TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments". Bioinformatics. 20 (11): 1806–1807. doi:10.1093/bioinformatics/bth155. PMID 14988107.
- ^ Etherington, Graham J.; Dicks, Jo; Roberts, Ian N. (1 February 2005). "Recombination Analysis Tool (RAT): a program for the high-throughput detection of recombination". Bioinformatics. 21 (3): 278–281. doi:10.1093/bioinformatics/bth500. PMID 15333462.
